General Statistics Examples
The technique of estimating parameters in linear models by using regression quantiles is a generalization of the LAE or LAV
least absolute value estimation technique. For a given quantile q, the estimate ![]() of
of ![]() in the model
in the model
is the value of b that minimizes
where ![]() and
and ![]() . For q = 0.5, the solution
. For q = 0.5, the solution ![]() is identical to the estimates that are produced by the LAE. The following routine finds this estimate by using linear programming.
is identical to the estimates that are produced by the LAE. The following routine finds this estimate by using linear programming.
This subroutine follows the approach given in Koenker and Bassett (1978); Bassett and Koenker (1982). When q = 0.5, this is equivalent to minimizing the sum of the absolute deviations, which is also known as L1 regression. For L1 regression, a faster and more accurate algorithm is available in the SAS/IML LAV subroutine, which is based on the approach given in Madsen and Nielsen (1993). For more information about quantile regression, see the documentation for the QUANTREG procedure in SAS/STAT User's Guide.
proc iml;
/*---------------------------------------------------------*/
/* Routine to find regression quantiles */
/* yname: name of dependent variable */
/* y: dependent variable */
/* xname: names of independent variables */
/* X: independent variables */
/* b: estimates */
/* predict: predicted values */
/* error: difference of y and predicted. */
/* q: quantile */
/*---------------------------------------------------------*/
start rq( yname, y, xname, X, b, predict, error, q);
bound=1.0e10;
coef = X`;
m = nrow(coef);
n = ncol(coef);
/*-----------------build rhs and bounds--------------------*/
e = repeat(1,1,n)`;
r = {0 0} || ((1-q)*coef*e)`;
sign = repeat(1,1,m);
do i=1 to m;
if r[2+i] < 0 then do;
sign[i] = -1;
r[2+i] = -r[2+i];
coef[i,] = -coef[i,];
end;
end;
l = repeat(0,1,n) || repeat(0,1,m) || -bound || -bound ;
u = repeat(1,1,n) || repeat(0,1,m) || { . . } ;
/*-------------build coefficient matrix and basis----------*/
a = ( y` || repeat(0,1,m) || { -1 0 } ) //
( repeat(0,1,n) || repeat(-1,1,m) || { 0 -1 } ) //
( coef || I(m) || repeat(0,m,2)) ;
/*----------------find the optimal solution----------------*/
cost = j(1,n+m+2,0);
cost[n+m+1] = 1.0;
call lpsolve(rc,optimum,p,d,rcost,
cost,a,r,-1,j(m+2,1,'E'),,l,u);
/*---------------- report the solution-----------------------*/
variable = xname`; b=d[3:m+2];
do i=1 to m;
b[i] = b[i] * sign[i];
end;
predict = X*b;
error = y - predict;
wsum = sum ( choose(error<0 , (q-1)*error , q*error) );
label = 'Estimation for ' + yname;
desc = q // n // wsum;
rownames = {'Regression Quantile', 'Number of Observations',
'Sum of Weighted Absolute Errors'};
print desc[L=label r=rownames];
print b[r=variable];
print X y predict error;
finish rq;
The following example uses data on the United States population from 1790 to 1970, and compares the L1 residuals to the least square residuals:
z = { 3.929 1790 ,
5.308 1800 ,
7.239 1810 ,
9.638 1820 ,
12.866 1830 ,
17.069 1840 ,
23.191 1850 ,
31.443 1860 ,
39.818 1870 ,
50.155 1880 ,
62.947 1890 ,
75.994 1900 ,
91.972 1910 ,
105.710 1920 ,
122.775 1930 ,
131.669 1940 ,
151.325 1950 ,
179.323 1960 ,
203.211 1970 };
y=z[,1];
x=repeat(1,19,1)||z[,2]||z[,2]##2;
run rq('pop',y,{'intercpt' 'year' 'yearsq'},x,b1,pred,resid,.5);
/* Compare L1 residuals with least squares residuals */
/* Compute the least squares residuals */
LSResid=y-x*inv(x`*x)*x`*y;
L1Resid = resid;
t = z[,2];
create Residuals var{t L1Resid LSResid}; append; close Residuals;
quit;
proc sgplot data=Residuals;
scatter x=t y=L1Resid / LEGENDLABEL= "L(1) residuals";
scatter x=t y=LSResid / LEGENDLABEL= "Least squares residuals";
yaxis label="Residuals";
refline 0 / axis=y;
run;
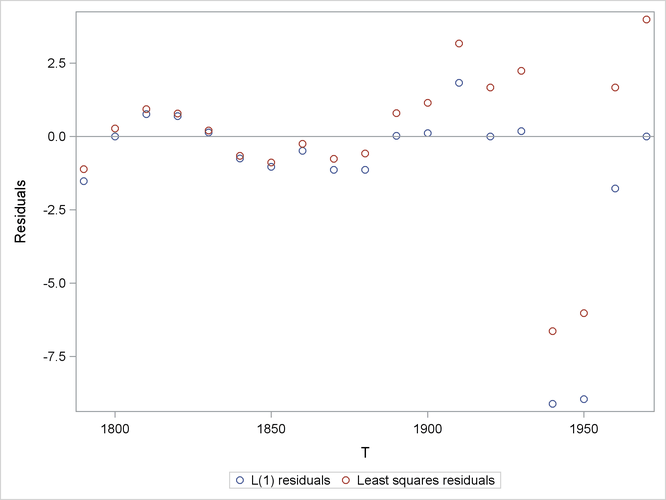
The results are shown in Output 9.11.1.
Output 9.11.1: Regression Quantiles: Results
| X | y | predict | error | ||
|---|---|---|---|---|---|
| 1 | 1790 | 3204100 | 3.929 | 5.4549176 | -1.525918 |
| 1 | 1800 | 3240000 | 5.308 | 5.308 | -9.02E-13 |
| 1 | 1810 | 3276100 | 7.239 | 6.4708902 | 0.7681098 |
| 1 | 1820 | 3312400 | 9.638 | 8.9435882 | 0.6944118 |
| 1 | 1830 | 3348900 | 12.866 | 12.726094 | 0.1399059 |
| 1 | 1840 | 3385600 | 17.069 | 17.818408 | -0.749408 |
| 1 | 1850 | 3422500 | 23.191 | 24.220529 | -1.029529 |
| 1 | 1860 | 3459600 | 31.443 | 31.932459 | -0.489459 |
| 1 | 1870 | 3496900 | 39.818 | 40.954196 | -1.136196 |
| 1 | 1880 | 3534400 | 50.155 | 51.285741 | -1.130741 |
| 1 | 1890 | 3572100 | 62.947 | 62.927094 | 0.0199059 |
| 1 | 1900 | 3610000 | 75.994 | 75.878255 | 0.1157451 |
| 1 | 1910 | 3648100 | 91.972 | 90.139224 | 1.8327765 |
| 1 | 1920 | 3686400 | 105.71 | 105.71 | 4.505E-12 |
| 1 | 1930 | 3724900 | 122.775 | 122.59058 | 0.1844157 |
| 1 | 1940 | 3763600 | 131.669 | 140.78098 | -9.111976 |
| 1 | 1950 | 3802500 | 151.325 | 160.28118 | -8.956176 |
| 1 | 1960 | 3841600 | 179.323 | 181.09118 | -1.768184 |
| 1 | 1970 | 3880900 | 203.211 | 203.211 | 6.821E-13 |
The L1 norm (when q = 0.5) tends to cause the fit to be better at more points at the expense of causing the fit to be worse at some points, as shown in Output 9.11.2, which shows a plot that compares the L1 residuals with the least squares residuals.
When q =0.5, the results of this module can be compared with the results of the LAV routine, as follows:
b0 = {1 1 1}; /* initial value */
optn = j(4,1,.); /* options vector */
optn[1]= .; /* gamma default */
optn[2]= 5; /* print all */
optn[3]= 0; /* McKean-Schradar variance */
optn[4]= 1; /* convergence test */
call LAV(rc, xr, x, y, b0, optn);