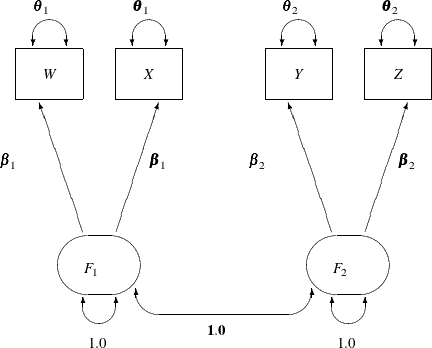
The path diagram for the one-factor model with parallel tests is shown in Figure 17.26.
The hypothesis ![]() differs from
differs from ![]() in that
in that F1 and F2 have a perfect correlation in ![]() . This is indicated by the fixed value 1.0 for the double-headed path that connects
. This is indicated by the fixed value 1.0 for the double-headed path that connects F1 and F2 in Figure 17.26. Again, you need only minimal modification of the preceding specification for ![]() to specify the path diagram in Figure 17.26, as shown in the following statements:
to specify the path diagram in Figure 17.26, as shown in the following statements:
proc calis data=lord;
path
W <=== F1 = beta1,
X <=== F1 = beta1,
Y <=== F2 = beta2,
Z <=== F2 = beta2;
pvar
F1 = 1.0,
F2 = 1.0,
W X = 2 * theta1,
Y Z = 2 * theta2;
pcov
F1 F2 = 1.0;
run;
The only modification of the preceding specification is in the PCOV statement, where you put a constant 1 for the covariance
between F1 and F2. An annotated fit summary is displayed in Figure 17.27.
Figure 17.27: Fit Summary, H1: One-Factor Model with Parallel Tests for Lord Data
| Fit Summary | |
|---|---|
| Chi-Square | 37.3337 |
| Chi-Square DF | 6 |
| Pr > Chi-Square | <.0001 |
| Standardized RMR (SRMR) | 0.0286 |
| Adjusted GFI (AGFI) | 0.9509 |
| RMSEA Estimate | 0.0898 |
| Bentler Comparative Fit Index | 0.9785 |
The chi-square value is 37.3337 (df=6, p<0.0001). This indicates that you can reject the hypothesized model H1 at the 0.01 ![]() -level. The standardized root mean square error (SRMSR) is 0.0286, the adjusted GFI (AGFI) is 0.9509, and Bentler’s comparative
fit index is 0.9785. All these indicate good model fit. However, the RMSEA is 0.0898, which does not support an acceptable
model for the data.
-level. The standardized root mean square error (SRMSR) is 0.0286, the adjusted GFI (AGFI) is 0.9509, and Bentler’s comparative
fit index is 0.9785. All these indicate good model fit. However, the RMSEA is 0.0898, which does not support an acceptable
model for the data.
The estimation results are displayed in Figure 17.28.
Figure 17.28: Estimation Results, H1: One-Factor Model with Parallel Tests for Lord Data
| PATH List | ||||||
|---|---|---|---|---|---|---|
| Path | Parameter | Estimate | Standard Error |
t Value | ||
| W | <=== | F1 | beta1 | 7.18623 | 0.26598 | 27.01802 |
| X | <=== | F1 | beta1 | 7.18623 | 0.26598 | 27.01802 |
| Y | <=== | F2 | beta2 | 8.44198 | 0.28000 | 30.14943 |
| Z | <=== | F2 | beta2 | 8.44198 | 0.28000 | 30.14943 |
| Variance Parameters | |||||
|---|---|---|---|---|---|
| Variance Type |
Variable | Parameter | Estimate | Standard Error |
t Value |
| Exogenous | F1 | 1.00000 | |||
| F2 | 1.00000 | ||||
| Error | W | theta1 | 34.68865 | 1.64634 | 21.07010 |
| X | theta1 | 34.68865 | 1.64634 | 21.07010 | |
| Y | theta2 | 26.28513 | 1.39955 | 18.78119 | |
| Z | theta2 | 26.28513 | 1.39955 | 18.78119 | |
| Covariances Among Exogenous Variables | ||||
|---|---|---|---|---|
| Var1 | Var2 | Estimate | Standard Error |
t Value |
| F1 | F2 | 1.00000 | ||
The goodness-of-fit tests for the four hypotheses are summarized in the following table.
|
Number of |
Degrees of |
||||
|---|---|---|---|---|---|
|
Hypothesis |
Parameters |
|
Freedom |
p-value |
|
|
|
4 |
37.33 |
6 |
< .0001 |
1.0 |
|
|
5 |
1.93 |
5 |
0.8583 |
0.8986 |
|
|
8 |
36.21 |
2 |
< .0001 |
1.0 |
|
|
9 |
0.70 |
1 |
0.4018 |
0.8986 |
Recall that the estimates of ![]() for
for ![]() and
and ![]() are almost identical, about 0.90, indicating that the speeded and unspeeded tests are measuring almost the same latent variable.
However, when
are almost identical, about 0.90, indicating that the speeded and unspeeded tests are measuring almost the same latent variable.
However, when ![]() was set to 1 in
was set to 1 in ![]() and
and ![]() (both one-factor models), both hypotheses were rejected. Hypotheses
(both one-factor models), both hypotheses were rejected. Hypotheses ![]() and
and ![]() (both two-factor models) seem to be consistent with the data. Since
(both two-factor models) seem to be consistent with the data. Since ![]() is obtained by adding four constraints (for the requirement of parallel tests) to
is obtained by adding four constraints (for the requirement of parallel tests) to ![]() (the full model), you can test
(the full model), you can test ![]() versus
versus ![]() by computing the differences of the chi-square statistics and their degrees of freedom, yielding a chi-square of 1.23 with
four degrees of freedom, which is obviously not significant. In a sense, the chi-square difference test means that representing
the data by
by computing the differences of the chi-square statistics and their degrees of freedom, yielding a chi-square of 1.23 with
four degrees of freedom, which is obviously not significant. In a sense, the chi-square difference test means that representing
the data by ![]() would not be significantly worse than representing the data by
would not be significantly worse than representing the data by ![]() . In addition, because
. In addition, because ![]() offers a more precise description of the data (with the assumption of parallel tests) than
offers a more precise description of the data (with the assumption of parallel tests) than ![]() , it should be chosen because of its simplicity. In conclusion, the two-factor model with parallel tests provides the best
explanation of the data.
, it should be chosen because of its simplicity. In conclusion, the two-factor model with parallel tests provides the best
explanation of the data.