-
DATA=SAS-data-set
-
specifies the SAS data set containing the data to be analyzed. If you omit the DATA= option, the procedure uses the most recently created SAS data set.
-
DESCENDING
DESCEND
DESC
-
specifies that the levels of the response variable for the ordinal multinomial model and the binomial model with single variable
response syntax be sorted in the reverse of the default order. For example, if RORDER=FORMATTED (the default), the DESCENDING
option causes the levels to be sorted from highest to lowest instead of from lowest to highest. If RORDER=FREQ, the DESCENDING
option causes the levels to be sorted from lowest frequency count to highest instead of from highest to lowest.
-
EXACTONLY
-
requests only the exact analyses. The asymptotic analysis that PROC GENMOD usually performs is suppressed.
-
NAMELEN=n
-
specifies the length of effect names in tables and output data sets to be n characters long, where n is a value between 20 and 200 characters. The default length is 20 characters.
-
ORDER=DATA | FORMATTED | FREQ | INTERNAL
-
specifies the sort order for the levels of the classification variables (which are specified in the CLASS statement). The ORDER= option can be useful when you use the CONTRAST or ESTIMATE statement because it determines which parameters in
the model correspond to each level in the data.
This option applies to the levels for all classification variables, except when you use the (default) ORDER=FORMATTED option
with numeric classification variables that have no explicit format. With this option, the levels of such variables are ordered
by their internal value.
The ORDER= option can take the following values:
|
Value of ORDER=
|
Levels Sorted By
|
|
DATA
|
Order of appearance in the input data set
|
|
FORMATTED
|
External formatted value, except for numeric variables with no explicit format, which are sorted by their unformatted (internal)
value
|
|
FREQ
|
Descending frequency count; levels with the most observations come first in the order
|
|
INTERNAL
|
Unformatted value
|
By default, ORDER=FORMATTED. For ORDER=FORMATTED and ORDER=INTERNAL, the sort order is machine-dependent. For more information
about sort order, see the chapter on the SORT procedure in the
Base SAS Procedures Guide and the discussion of BY-group processing in
SAS Language Reference: Concepts.
-
PLOTS <(global-plot-option)>= plot-request <(options)>
PLOTS <(global-plot-options)> <= (plot-request <(options)> <…plot-request <(options)>>)>
-
specifies plots to be created using ODS Graphics. Many of the observational statistics in the output data set can be plotted using this option. You are not required to create an output data set in order
to produce a plot. When you specify only one plot request, you can omit the parentheses around the plot request. Here are
some examples:
plots=all
plots=predicted
plots=(predicted reschi)
plots(unpack)=dfbeta
ODS Graphics must be enabled before plots can be requested. For example:
proc genmod plots=all;
model y = x;
run;
For more information about enabling and disabling ODS Graphics, see the section Enabling and Disabling ODS Graphics in Chapter 21: Statistical Graphics Using ODS.
Any specified global plot options apply to all plots that are specified with plot requests. The following global plot options
are available.
-
CLUSTERLABEL
-
displays formatted levels of the SUBJECT= effect instead of plot symbols. This option applies only to diagnostic statistics
for models fit by GEEs that are plotted against cluster number, and provides a way to identify cluster level names with corresponding
ordered cluster numbers.
-
UNPACK
-
displays multiple plots individually. The default is to display related multiple plots in a panel.
See the section OUTPUT Statement for definitions of the statistics specified with the plot requests. The plot requests include the following:
-
ALL
-
produces all available plots.
-
COOKSD
DOBS
-
plots the Cook’s distance statistic as a function of observation number.
-
DFBETA
-
plots the 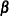 deletion statistic as a function of observation number for each regression parameter in the model.
deletion statistic as a function of observation number for each regression parameter in the model.
-
DFBETAS
-
plots the standardized 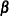 deletion statistic as a function of observation number for each regression parameter in the model.
deletion statistic as a function of observation number for each regression parameter in the model.
-
LEVERAGE
-
plots the leverage as a function of observation number.
-
PREDICTED<(option)>
-
plots predicted values with confidence limits as a function of observation number. The PREDICTED plot request has the following
option:
-
CLM
-
includes confidence limits in the predicted value plot.
-
PZERO
-
plots the zero inflation probability for zero-inflated Poisson and negative binomial models as a function of observation number.
-
RESCHI<(options)>
-
The RESCHI plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, Pearson residuals are plotted as a function of observation number.
-
RESDEV<(options)>
-
plots deviance residuals. The RESDEV plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, deviance residuals are plotted as a function of observation number.
-
RESLIK<(options)>
-
plots likelihood residuals. The RESLIK plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, likelihood residuals are plotted as a function of observation number.
-
RESRAW<(options)>
-
plots raw residuals. The RESRAW plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, raw residuals are plotted as a function of observation number.
-
STDRESCHI<(options)>
-
plots standardized Pearson residuals. The STDRESCHI plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, standardized Pearson residuals are plotted as a function of observation number.
-
STDRESDEV<(options)>
-
plots standardized deviance residuals. The STDRESDEV plot request has the following options:
-
INDEX
-
plots as a function of observation number.
-
XBETA
-
plots as a function of linear predictor.
If you do not specify an option, standardized deviance residuals are plotted as a function of observation number.
If you fit a model by using generalized estimating equations (GEEs), the following additional plot requests are available:
-
CLEVERAGE
-
plots the cluster leverage as a function of ordered cluster.
-
CLUSTERCOOKSD
DCLS
-
plots the cluster Cook’s distance statistic as a function of ordered cluster.
-
CLUSTERDFIT
MCLS
-
plots the studentized cluster Cook’s distance statistic as a function of ordered cluster.
-
DFBETAC
-
plots the cluster deletion statistic as a function of ordered cluster for each regression parameter in the model.
-
DFBETACS
-
plots the standardized cluster deletion statistic as a function of ordered cluster for each regression parameter in the model.
-
RORDER=keyword
-
specifies the sort order for the levels of the response variable. This order determines which intercept parameter in the model corresponds to each level in the data. If RORDER=FORMATTED for numeric variables
for which you have supplied no explicit format, the levels are ordered by their internal values. The following table displays
the valid keywords and describes how PROC GENMOD interprets them.
|
RORDER=keyword
|
Levels Sorted by
|
|
DATA
|
Order of appearance in the input data set
|
|
FORMATTED
|
External formatted value, except for numeric
|
| |
variables with no explicit format, which are
|
| |
sorted by their unformatted (internal) value
|
|
FREQ
|
Descending frequency count; levels with the
|
| |
most observations come first in the order
|
|
INTERNAL
|
Unformatted value
|
By default, RORDER=FORMATTED. For RORDER=FORMATTED and RORDER=INTERNAL, the sort order is machine dependent. The DESCENDING
option in the PROC GENMOD statement causes the response variable to be sorted in the reverse of the order displayed in the
previous table. For more information about sort order, see the chapter on the SORT procedure in the Base SAS Procedures Guide.
The NOPRINT option, which suppresses displayed output in other SAS procedures, is not available in the PROC GENMOD statement.
However, you can use the Output Delivery System (ODS) to suppress all displayed output, store all output on disk for further analysis, or create SAS data sets from selected output. You can suppress all displayed
output with the statement ODS SELECT NONE; and turn displayed output back on with the statement ODS SELECT ALL;. See Table 40.12 and Table 40.13 for the names of output tables available from PROC GENMOD. For more information about ODS, see Chapter 20: Using the Output Delivery System.


 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression