The MIANALYZE Procedure
- Overview
- Getting Started
-
Syntax

-
Details

-
Examples
 Reading Means and Standard Errors from a DATA= Data SetReading Means and Covariance Matrices from a DATA= COV Data SetReading Regression Results from a DATA= EST Data SetReading Mixed Model Results from PARMS= and COVB= Data SetsReading Generalized Linear Model ResultsReading GLM Results from PARMS= and XPXI= Data SetsReading Logistic Model Results from a PARMS= Data SetReading Mixed Model Results with Classification CovariatesReading Nominal Logistic Model ResultsUsing a TEST statementCombining Correlation CoefficientsSensitivity Analysis with Control-Based Pattern ImputationSensitivity Analysis with Tipping-Point Approach
Reading Means and Standard Errors from a DATA= Data SetReading Means and Covariance Matrices from a DATA= COV Data SetReading Regression Results from a DATA= EST Data SetReading Mixed Model Results from PARMS= and COVB= Data SetsReading Generalized Linear Model ResultsReading GLM Results from PARMS= and XPXI= Data SetsReading Logistic Model Results from a PARMS= Data SetReading Mixed Model Results with Classification CovariatesReading Nominal Logistic Model ResultsUsing a TEST statementCombining Correlation CoefficientsSensitivity Analysis with Control-Based Pattern ImputationSensitivity Analysis with Tipping-Point Approach - References
You specify input data sets based on the type of inference you requested. For univariate inference, you can use one of the following options:
-
a DATA= data set, which provides both parameter estimates and the associated standard errors
-
a DATA=EST, COV, or CORR data set, which provides both parameter estimates and the associated standard errors either explicitly (type CORR) or through the covariance matrix (type EST, COV)
-
PARMS= data set, which provides both parameter estimates and the associated standard errors
For multivariate inference, which includes the testing of linear hypotheses about parameters, you can use one of the following option combinations:
-
a DATA=EST, COV, or CORR data set, which provides parameter estimates and the associated covariance matrix either explicitly (type EST, COV) or through the correlation matrix and standard errors (type CORR) in a single data set
-
PARMS= and COVB= data sets, which provide parameter estimates in a PARMS= data set and the associated covariance matrix in a COVB= data set
-
PARMS=, COVB=, and PARMINFO= data sets, which provide parameter estimates in a PARMS= data set, the associated covariance matrix in a COVB= data set with variables named
PRM1,PRM2, …, and the effects associated with these variables in a PARMINFO= data set -
PARMS= and XPXI= data sets, which provide parameter estimates and the associated standard errors in a PARMS= data set and the associated
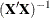 matrix in an XPXI= data set
matrix in an XPXI= data set
The appropriate combination depends on the type of inference and the SAS procedure you used to create the data sets. For instance, if you used PROC REG to create an OUTEST= data set that contains the parameter estimates and covariance matrix, you would use the DATA= option to read the OUTEST= data set.
When the input DATA= data set is a specially structured SAS data set, the data set must contain the variable _Imputation_ to identify the imputation by number. Otherwise, each observation corresponds to an imputation and contains both parameter
estimates and associated standard errors.
If you do not specify an input data set with the DATA= or PARMS= option, then the most recently created SAS data set is used as an input DATA= data set. Note that with a DATA= data set, each effect represents a continuous variable; only regressor effects (continuous variables by themselves) are allowed in the MODELEFFECTS statement.
The DATA= data set provides both parameter estimates and the associated standard errors computed from imputed data sets. Such data sets are typically created with an OUTPUT statement in procedures such as PROC MEANS and PROC UNIVARIATE.
The MIANALYZE procedure reads parameter estimates from observations with variables in the MODELEFFECTS statement, and standard errors for parameter estimates from observations with variables in the STDERR statement. The order of the variables for standard errors must match the order of the variables for parameter estimates.
The specially structured DATA= data set provides both parameter estimates and the associated covariance matrix computed from imputed data sets. Such data sets are created by procedures such as PROC CORR (type COV, CORR) and PROC REG (type EST).
With a DATA=EST data set, the MIANALYZE procedure reads parameter estimates from observations with _TYPE_=‘PARM’, _TYPE_=‘PARMS’, _TYPE_=‘OLS’, or _TYPE_=‘FINAL’, and covariance matrices for parameter estimates from observations with _TYPE_=‘COV’ or _TYPE_=‘COVB’.
With a DATA=COV data set, the procedure reads sample means from observations with _TYPE_=‘MEAN’, sample size n from observations with _TYPE_=‘N’, and covariance matrices for variables from observations with _TYPE_=‘COV’.
With a DATA=CORR data set, the procedure reads sample means from observations with _TYPE_=‘MEAN’, sample size n from observations with _TYPE_=‘N’, correlation matrices for variables from observations with _TYPE_=‘CORR’, and standard errors for variables from observations with _TYPE_=‘STD’. The standard errors and correlation matrix are used to generate a covariance matrix for the variables.
Note that with a DATA=COV or DATA=CORR data set, each covariance matrix for the variables is divided by n to create the covariance matrix for the sample means.
The PARMS= data set contains both parameter estimates and the associated standard errors computed from imputed data sets. Such data sets are typically created with an ODS OUTPUT statement in procedures such as PROC GENMOD, PROC GLM, PROC LOGISTIC, and PROC MIXED.
The MIANALYZE procedure reads effect names from observations with the variable Parameter, Effect, Variable, or Parm. It then reads parameter estimates from observations with the variable Estimate and standard errors for parameter estimates from observations with the variable StdErr.
The available parms-options include the CLASSVAR= option to identify classification variables and the LINK= option to input logistic regression results. When the parameter estimates are derived from the LOGISTIC procedure, the LINK= option can be used to identify the variable required when the parameter estimates are read from observations. The available options are as follows:
-
LINK=NONE (which is the default), in which each model effect is completely identified from the effect name. This option should be used for all procedures except PROC LOGISTIC.
-
LINK=LOGIT, in which the variable
ClassVal0is used to identify response levels forInterceptfrom PROC LOGISTIC for ordinal responses. This option is applicable only when the variableInterceptis in the MODELEFFECTS statement and the logistic model has more than two response levels. Otherwise, LINK=NONE should be used. -
LINK=GLOGIT, in which the variable
Responseis used to identify response levels for the parameters from PROC LOGISTIC for nominal responses.
When the effects contain classification variables, the CLASSVAR= option can be used to identify the variables when reading the classification levels from observations. The available options are:
-
CLASSVAR=FULL (which is the default), the data set contains the classification variables explicitly. PROC MIANALYZE reads the classification levels from observations with their corresponding classification variables. PROC MIXED generates this type of table.
-
CLASSVAR=LEVEL, PROC MIANALYZE reads the classification levels for the effect from observations with variables
Level1,Level2, and so on, where the variableLevel1contains the classification level for the first classification variable in the effect, and the variableLevel2contains the classification level for the second classification variable in the effect. For each effect, the variables in the crossed list are displayed before the variables in the nested list. The variable order in the CLASS statement is used for variables inside each list. PROC GENMOD generates this type of table.For example, with the following statements, the variable
Level1has the classification level of the variablec2for the effectc2:proc mianalyze parms(classvar=Level)=dataparm; class c1 c2 c3; modeleffects c2 c3(c2 c1); run;
For the effect
c3(c2 c1), the variableLevel1has the classification level of the variablec3,Level2has the level ofc1, andLevel3has the level ofc2. -
CLASSVAR=CLASSVAL, PROC MIANALYZE reads the classification levels for the effect from observations with variables
ClassVal0,ClassVal1, and so on, where the variableClassVal0contains the classification level for the first classification variable in the effect, and the variableClassVal1contains the classification level for the second classification variable in the effect. For each effect, the variables in the crossed list are displayed before the variables in the nested list. The variable order in the CLASS statement is used for variables inside each list. PROC LOGISTIC generates this type of tables.
The PARMS= data set contains parameter estimates, and the COVB= data set contains associated covariance matrices computed from imputed data sets. Such data sets are typically created with an ODS OUTPUT statement in procedures such as PROC LOGISTIC, PROC MIXED, and PROC REG.
When you specify a PARMS= data set, the MIANALYZE procedure reads effect names from observations with the variable Parameter, Effect, Variable, or Parm. It then reads parameter estimates from observations with the variable Estimate.
The available parms-options include the CLASSVAR= option to identify classification variables and the LINK= option to input logistic regression results. For a detailed description of the PARMS= option, see the section PARMS <( parms-options)>= Data Set.
The EFFECTVAR=etype option identifies the variables for parameters displayed in the covariance matrix. The available types are STACKING and ROWCOL:
-
EFFECTVAR=STACKING (which is the default), each parameter is displayed by stacking variables in the effect. Begin with the variables in the crossed list, followed by the continuous list, then followed by the nested list. Each classification variable is displayed with its classification level attached. PROC LOGISTIC generates this type of table. When each effect is a continuous variable by itself, each stacked parameter name reduces to the effect name. PROC REG generates this type of table.
The MIANALYZE procedure reads parameter names from observations with the variable
Parameter,Effect,Variable,Parm, orRowName. It then reads covariance matrices from observations with the stacked variables in a COVB= data set. -
EFFECTVAR=ROWCOL, parameters are displayed by the variables
Col1,Col2, ... The parameter associated with the variableCol1is identified by the observation with value 1 for the variableRow. The parameter associated with the variableCol2is identified by the observation with value 2 for the variableRow. PROC MIXED generates this type of table.The MIANALYZE procedure reads the parameter indices from observations with the variable
Rowand the effect names from observations with the variableParameter,Effect,Variable,Parm, orRowName. It then reads covariance matrices from observations with the variablesCol1,Col2, and so on in a COVB= data set.When the effects contain classification variables, the data set contains the classification variables explicitly and the MIANALYZE procedure also reads the classification levels from their corresponding classification variables.
The input PARMS= data set contains parameter estimates, the PARMINFO= data set identifies parameters with the variables Prm1, Prm2, and so on, and the COVB= data set contains associated covariance matrices with the variables Prm1, Prm2, and so on. Such data sets are typically created with an ODS OUTPUT statement using procedure such as PROC GENMOD.
When you specify a PARMS= data set, the MIANALYZE procedure reads effect names from observations with the variable Parameter, Effect, Variable, or Parm. It then reads parameter estimates from observations with the variable Estimate.
When the effects contain classification variables, the option CLASSVAR= ctype can be used to identify the associated classification variables when reading the classification levels from observations. The available types are FULL, LEVEL, and CLASSVAL, and they are described in the section PARMS <( parms-options)>= Data Set. The default is CLASSVAR= FULL.
When you specify a COVB= data set, the MIANALYZE procedure reads parameter names from observations with the variable Parameter, Effect, Variable, Parm, or RowName. It then reads covariance matrices from observations with the variables Prm1, Prm2, and so on.
The parameters associated with the variables Prm1, Prm2, and so on are identified in the PARMINFO= data set. PROC MIANALYZE reads the parameter names from observations with the
variable Parameter and the corresponding effect from observations with the variable Effect. When the effects contain classification variables, the data set contains the classification variables explicitly and the
MIANALYZE procedure also reads the classification levels from observations with their corresponding classification variables.
The input PARMS= data set contains parameter estimates, and the input XPXI= data set contains associated ![]() matrices computed from imputed data sets. Such data sets are typically created with an ODS OUTPUT statement in a procedure
such as PROC GLM.
matrices computed from imputed data sets. Such data sets are typically created with an ODS OUTPUT statement in a procedure
such as PROC GLM.
When you specify a PARMS= data set, the MIANALYZE procedure reads parameter names from observations with the variable Parameter, Effect, Variable, or Parm. It then reads parameter estimates from observations with the variable Estimate and standard errors for parameter estimates from observations with the variable StdErr.
When you specify a XPXI= data set, the MIANALYZE procedure reads parameter names from observations with the variable Parameter and ![]() matrices from observations with the parameter variables in the data set.
matrices from observations with the parameter variables in the data set.
Note that this combination can be used only when each effect is a continuous variable by itself.