The GLM Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 Statistical Assumptions for Using PROC GLMSpecification of EffectsUsing PROC GLM InteractivelyParameterization of PROC GLM ModelsHypothesis Testing in PROC GLMEffect Size Measures for F Tests in GLMAbsorptionSpecification of ESTIMATE ExpressionsComparing GroupsMultivariate Analysis of VarianceRepeated Measures Analysis of VarianceRandom-Effects AnalysisMissing ValuesComputational ResourcesComputational MethodOutput Data SetsDisplayed OutputODS Table NamesODS Graphics
Statistical Assumptions for Using PROC GLMSpecification of EffectsUsing PROC GLM InteractivelyParameterization of PROC GLM ModelsHypothesis Testing in PROC GLMEffect Size Measures for F Tests in GLMAbsorptionSpecification of ESTIMATE ExpressionsComparing GroupsMultivariate Analysis of VarianceRepeated Measures Analysis of VarianceRandom-Effects AnalysisMissing ValuesComputational ResourcesComputational MethodOutput Data SetsDisplayed OutputODS Table NamesODS Graphics -
Examples
 Randomized Complete Blocks with Means Comparisons and ContrastsRegression with Mileage DataUnbalanced ANOVA for Two-Way Design with InteractionAnalysis of CovarianceThree-Way Analysis of Variance with ContrastsMultivariate Analysis of VarianceRepeated Measures Analysis of VarianceMixed Model Analysis of Variance with the RANDOM StatementAnalyzing a Doubly Multivariate Repeated Measures DesignTesting for Equal Group VariancesAnalysis of a Screening Design
Randomized Complete Blocks with Means Comparisons and ContrastsRegression with Mileage DataUnbalanced ANOVA for Two-Way Design with InteractionAnalysis of CovarianceThree-Way Analysis of Variance with ContrastsMultivariate Analysis of VarianceRepeated Measures Analysis of VarianceMixed Model Analysis of Variance with the RANDOM StatementAnalyzing a Doubly Multivariate Repeated Measures DesignTesting for Equal Group VariancesAnalysis of a Screening Design - References
Analysis of covariance combines some of the features of both regression and analysis of variance. Typically, a continuous variable (the covariate) is introduced into the model of an analysis-of-variance experiment.
Data in the following example are selected from a larger experiment on the use of drugs in the treatment of leprosy (Snedecor and Cochran, 1967, p. 422).
Variables in the study are as follows:
|
|
two antibiotics (A and D) and a control (F) |
|
|
a pretreatment score of leprosy bacilli |
|
|
a posttreatment score of leprosy bacilli |
Ten patients are selected for each treatment (Drug), and six sites on each patient are measured for leprosy bacilli.
The covariate (a pretreatment score) is included in the model for increased precision in determining the effect of drug treatments on the posttreatment count of bacilli.
The following statements create the data set, perform a parallel-slopes analysis of covariance with PROC GLM, and compute Drug LS-means. These statements produce Output 44.4.1 and Output 44.4.2.
data DrugTest; input Drug $ PreTreatment PostTreatment @@; datalines; A 11 6 A 8 0 A 5 2 A 14 8 A 19 11 A 6 4 A 10 13 A 6 1 A 11 8 A 3 0 D 6 0 D 6 2 D 7 3 D 8 1 D 18 18 D 8 4 D 19 14 D 8 9 D 5 1 D 15 9 F 16 13 F 13 10 F 11 18 F 9 5 F 21 23 F 16 12 F 12 5 F 12 16 F 7 1 F 12 20 ;
proc glm data=DrugTest; class Drug; model PostTreatment = Drug PreTreatment / solution; lsmeans Drug / stderr pdiff cov out=adjmeans; run;
proc print data=adjmeans; run;
Output 44.4.1: Classes and Levels
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Drug | 3 | A D F |
| Number of Observations Read | 30 |
|---|---|
| Number of Observations Used | 30 |
Output 44.4.2: Overall Analysis of Variance
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 3 | 871.497403 | 290.499134 | 18.10 | <.0001 |
| Error | 26 | 417.202597 | 16.046254 | ||
| Corrected Total | 29 | 1288.700000 |
| R-Square | Coeff Var | Root MSE | PostTreatment Mean |
|---|---|---|---|
| 0.676261 | 50.70604 | 4.005778 | 7.900000 |
This model assumes that the slopes relating posttreatment scores to pretreatment scores are parallel for all drugs. You can
check this assumption by including the class-by-covariate interaction, Drug*PreTreatment, in the model and examining the ANOVA test for the significance of this effect. This extra test is omitted in this example,
but it is insignificant, justifying the equal-slopes assumption.
In Output 44.4.3, the Type I SS for Drug (293.6) gives the between-drug sums of squares that are obtained for the analysis-of-variance model PostTreatment=Drug. This measures the difference between arithmetic means of posttreatment scores for different drugs, disregarding the covariate.
The Type III SS for Drug (68.5537) gives the Drug sum of squares adjusted for the covariate. This measures the differences between Drug LS-means, controlling for the covariate. The Type I test is highly significant (p = 0.001), but the Type III test is not. This indicates that, while there is a statistically significant difference between
the arithmetic drug means, this difference is reduced to below the level of background noise when you take the pretreatment
scores into account. From the table of parameter estimates, you can derive the least squares predictive formula model for
estimating posttreatment score based on pretreatment score and drug:
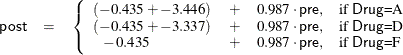
Output 44.4.3: Tests and Parameter Estimates
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Drug | 2 | 293.6000000 | 146.8000000 | 9.15 | 0.0010 |
| PreTreatment | 1 | 577.8974030 | 577.8974030 | 36.01 | <.0001 |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Drug | 2 | 68.5537106 | 34.2768553 | 2.14 | 0.1384 |
| PreTreatment | 1 | 577.8974030 | 577.8974030 | 36.01 | <.0001 |
| Parameter | Estimate | Standard Error | t Value | Pr > |t| | |
|---|---|---|---|---|---|
| Intercept | -0.434671164 | B | 2.47135356 | -0.18 | 0.8617 |
| Drug A | -3.446138280 | B | 1.88678065 | -1.83 | 0.0793 |
| Drug D | -3.337166948 | B | 1.85386642 | -1.80 | 0.0835 |
| Drug F | 0.000000000 | B | . | . | . |
| PreTreatment | 0.987183811 | 0.16449757 | 6.00 | <.0001 |
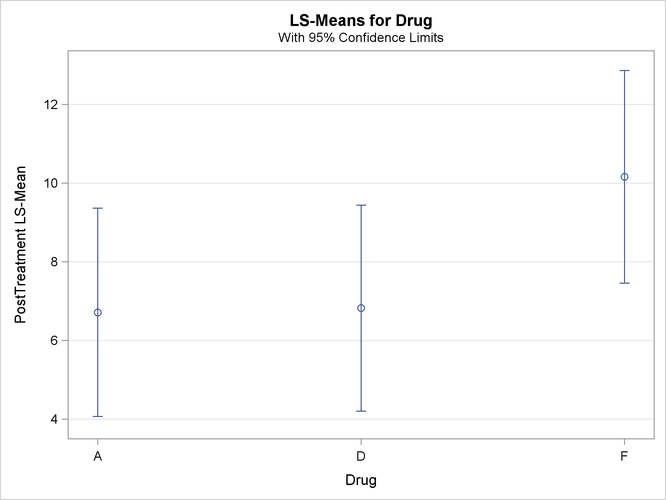
Output 44.4.4 displays the LS-means, which are, in a sense, the means adjusted for the covariate. The STDERR option in the LSMEANS statement causes the standard error of the LS-means and the probability of getting a larger t value under the hypothesis ![]() to be included in this table as well. Specifying the PDIFF option causes all probability values for the hypothesis
to be included in this table as well. Specifying the PDIFF option causes all probability values for the hypothesis ![]() to be displayed, where the indexes i and j are numbered treatment levels.
to be displayed, where the indexes i and j are numbered treatment levels.
Output 44.4.4: LS-Means
| Drug | PostTreatment LSMEAN | Standard Error | Pr > |t| | LSMEAN Number |
|---|---|---|---|---|
| A | 6.7149635 | 1.2884943 | <.0001 | 1 |
| D | 6.8239348 | 1.2724690 | <.0001 | 2 |
| F | 10.1611017 | 1.3159234 | <.0001 | 3 |
| Least Squares Means for effect Drug Pr > |t| for H0: LSMean(i)=LSMean(j) Dependent Variable: PostTreatment |
|||
|---|---|---|---|
| i/j | 1 | 2 | 3 |
| 1 | 0.9521 | 0.0793 | |
| 2 | 0.9521 | 0.0835 | |
| 3 | 0.0793 | 0.0835 | |
The OUT= and COV options in the LSMEANS statement create a data set of the estimates, their standard errors, and the variances and covariances of the LS-means, which is displayed in Output 44.4.5.
Output 44.4.5: LS-Means Output Data Set
| Obs | _NAME_ | Drug | LSMEAN | STDERR | NUMBER | COV1 | COV2 | COV3 |
|---|---|---|---|---|---|---|---|---|
| 1 | PostTreatment | A | 6.7150 | 1.28849 | 1 | 1.66022 | 0.02844 | -0.08403 |
| 2 | PostTreatment | D | 6.8239 | 1.27247 | 2 | 0.02844 | 1.61918 | -0.04299 |
| 3 | PostTreatment | F | 10.1611 | 1.31592 | 3 | -0.08403 | -0.04299 | 1.73165 |
The new graphical features of PROC GLM enable you to visualize the fitted analysis of covariance model. The following statements
enable ODS Graphics by specifying the ODS GRAPHICS statement and then fit an analysis-of-covariance model with LS-means for
Drug.
ods graphics on; proc glm data=DrugTest plot=meanplot(cl); class Drug; model PostTreatment = Drug PreTreatment; lsmeans Drug / pdiff; run; ods graphics off;
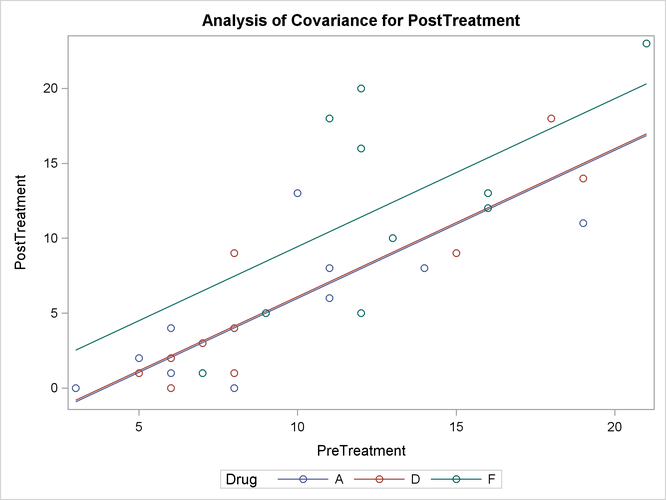
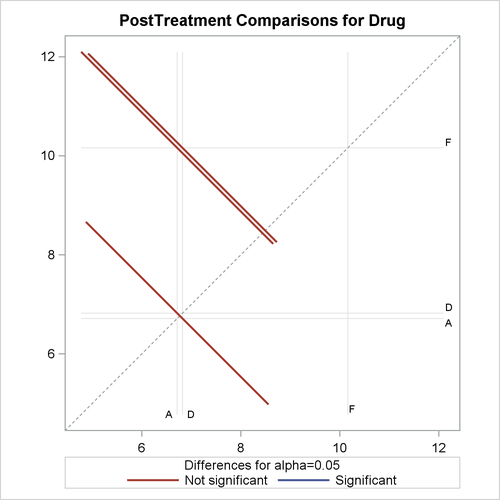
With graphics enabled, the GLM procedure output includes an analysis-of-covariance plot, as in Output 44.4.6. The LSMEANS statement produces a plot of the LS-means; the SAS statements previously shown use the PLOTS=MEANPLOT(CL) option to add confidence limits for the individual LS-means, shown in Output 44.4.7. If you also specify the PDIFF option in the LSMEANS statement, the output also includes a plot appropriate for the type of LS-mean differences computed. In this case, the default is to compare all LS-means with each other pairwise, so the plot is a “diffogram” or “mean-mean scatter plot” (Hsu, 1996), as in Output 44.4.8. For general information about ODS Graphics, see Chapter 21: Statistical Graphics Using ODS. For specific information about the graphics available in the GLM procedure, see the section ODS Graphics.
The analysis of covariance plot Output 44.4.6 makes it clear that the control (drug F) has higher posttreatment scores across the range of pretreatment scores, while the fitted models for the two antibiotics (drugs A and D) nearly coincide. Similarly, while the diffogram Output 44.4.7 indicates that none of the LS-mean differences are significant at the 5% level, the difference between the LS-means for the two antibiotics is much closer to zero than the differences between either one and the control.