The MI Procedure
- Overview
- Getting Started
-
Syntax

-
Details
 Descriptive StatisticsEM Algorithm for Data with Missing ValuesStatistical Assumptions for Multiple ImputationMissing Data PatternsImputation MethodsMonotone Methods for Data Sets with Monotone Missing PatternsMonotone and FCS Regression MethodsMonotone and FCS Predictive Mean Matching MethodsMonotone Propensity Score MethodMonotone and FCS Discriminant Function MethodsMonotone and FCS Logistic Regression MethodsFCS Methods for Data Sets with Arbitrary Missing PatternsChecking Convergence in FCS MethodsMCMC Method for Arbitrary Missing Multivariate Normal DataProducing Monotone Missingness with the MCMC MethodMCMC Method SpecificationsChecking Convergence in MCMCInput Data SetsOutput Data SetsCombining Inferences from Multiply Imputed Data SetsMultiple Imputation EfficiencyImputer’s Model Versus Analyst’s ModelParameter Simulation versus Multiple ImputationSummary of Issues in Multiple ImputationODS Table NamesODS Graphics
Descriptive StatisticsEM Algorithm for Data with Missing ValuesStatistical Assumptions for Multiple ImputationMissing Data PatternsImputation MethodsMonotone Methods for Data Sets with Monotone Missing PatternsMonotone and FCS Regression MethodsMonotone and FCS Predictive Mean Matching MethodsMonotone Propensity Score MethodMonotone and FCS Discriminant Function MethodsMonotone and FCS Logistic Regression MethodsFCS Methods for Data Sets with Arbitrary Missing PatternsChecking Convergence in FCS MethodsMCMC Method for Arbitrary Missing Multivariate Normal DataProducing Monotone Missingness with the MCMC MethodMCMC Method SpecificationsChecking Convergence in MCMCInput Data SetsOutput Data SetsCombining Inferences from Multiply Imputed Data SetsMultiple Imputation EfficiencyImputer’s Model Versus Analyst’s ModelParameter Simulation versus Multiple ImputationSummary of Issues in Multiple ImputationODS Table NamesODS Graphics -
Examples
 EM Algorithm for MLEMonotone Propensity Score MethodMonotone Regression MethodMonotone Logistic Regression Method for CLASS VariablesMonotone Discriminant Function Method for CLASS VariablesFCS Method for Continuous VariablesFCS Method for CLASS VariablesFCS Method with Trace PlotMCMC MethodProducing Monotone Missingness with MCMCChecking Convergence in MCMCSaving and Using Parameters for MCMCTransforming to NormalityMultistage Imputation
EM Algorithm for MLEMonotone Propensity Score MethodMonotone Regression MethodMonotone Logistic Regression Method for CLASS VariablesMonotone Discriminant Function Method for CLASS VariablesFCS Method for Continuous VariablesFCS Method for CLASS VariablesFCS Method with Trace PlotMCMC MethodProducing Monotone Missingness with MCMCChecking Convergence in MCMCSaving and Using Parameters for MCMCTransforming to NormalityMultistage Imputation - References
MCMC Method for Arbitrary Missing Multivariate Normal Data
The Markov chain Monte Carlo (MCMC) method originated in physics as a tool for exploring equilibrium distributions of interacting molecules. In statistical applications, it is used to generate pseudorandom draws from multidimensional and otherwise intractable probability distributions via Markov chains. A Markov chain is a sequence of random variables in which the distribution of each element depends only on the value of the previous element.
In MCMC simulation, you constructs a Markov chain long enough for the distribution of the elements to stabilize to a stationary distribution, which is the distribution of interest. Repeatedly simulating steps of the chain simulates draws from the distribution of interest. See Schafer (1997) for a detailed discussion of this method.
In Bayesian inference, information about unknown parameters is expressed in the form of a posterior probability distribution. This posterior distribution is computed using Bayes’ theorem,
|
|
MCMC has been applied as a method for exploring posterior distributions in Bayesian inference. That is, through MCMC, you can simulate the entire joint posterior distribution of the unknown quantities and obtain simulation-based estimates of posterior parameters that are of interest.
In many incomplete-data problems, the observed-data posterior ![]() is intractable and cannot easily be simulated. However, when
is intractable and cannot easily be simulated. However, when ![]() is augmented by an estimated or simulated value of the missing data
is augmented by an estimated or simulated value of the missing data ![]() , the complete-data posterior
, the complete-data posterior ![]() is much easier to simulate. Assuming that the data are from a multivariate normal distribution, data augmentation can be
applied to Bayesian inference with missing data by repeating the following steps:
is much easier to simulate. Assuming that the data are from a multivariate normal distribution, data augmentation can be
applied to Bayesian inference with missing data by repeating the following steps:
1. The imputation I-step Given an estimated mean vector and covariance matrix, the I-step simulates the missing values for each observation independently.
That is, if you denote the variables with missing values for observation i by ![]() and the variables with observed values by
and the variables with observed values by ![]() , then the I-step draws values for
, then the I-step draws values for ![]() from a conditional distribution for
from a conditional distribution for ![]() given
given ![]() .
.
2. The posterior P-step Given a complete sample, the P-step simulates the posterior population mean vector and covariance matrix. These new estimates are then used in the next I-step. Without prior information about the parameters, a noninformative prior is used. You can also use other informative priors. For example, a prior information about the covariance matrix can help to stabilize the inference about the mean vector for a near singular covariance matrix.
That is, with a current parameter estimate ![]() at the
at the ![]() iteration, the I-step draws
iteration, the I-step draws ![]() from
from ![]() and the P-step draws
and the P-step draws ![]() from
from ![]() . The two steps are iterated long enough for the results to reliably simulate an approximately independent draw of the missing
values for a multiply imputed data set (Schafer, 1997).
. The two steps are iterated long enough for the results to reliably simulate an approximately independent draw of the missing
values for a multiply imputed data set (Schafer, 1997).
This creates a Markov chain ![]() ,
, ![]() , …, which converges in distribution to
, …, which converges in distribution to ![]() . Assuming the iterates converge to a stationary distribution, the goal is to simulate an approximately independent draw of
the missing values from this distribution.
. Assuming the iterates converge to a stationary distribution, the goal is to simulate an approximately independent draw of
the missing values from this distribution.
To validate the imputation results, you should repeat the process with different random number generators and starting values based on different initial parameter estimates.
The next three sections provide details for the imputation step, Bayesian estimation of the mean vector and covariance matrix, and the posterior step.
Imputation Step
In each iteration, starting with a given mean vector ![]() and covariance matrix
and covariance matrix ![]() , the imputation step draws values for the missing data from the conditional distribution
, the imputation step draws values for the missing data from the conditional distribution ![]() given
given ![]() .
.
Suppose ![]() is the partitioned mean vector of two sets of variables,
is the partitioned mean vector of two sets of variables, ![]() and
and ![]() , where
, where ![]() is the mean vector for variables
is the mean vector for variables ![]() and
and ![]() is the mean vector for variables
is the mean vector for variables ![]() .
.
Also suppose
|
|
|
|
is the partitioned covariance matrix for these variables, where ![]() is the covariance matrix for variables
is the covariance matrix for variables ![]() ,
, ![]() is the covariance matrix for variables
is the covariance matrix for variables ![]() , and
, and ![]() is the covariance matrix between variables
is the covariance matrix between variables ![]() and variables
and variables ![]() .
.
By using the sweep operator (Goodnight, 1979) on the pivots of the ![]() submatrix, the matrix becomes
submatrix, the matrix becomes
|
|
where ![]() can be used to compute the conditional covariance matrix of
can be used to compute the conditional covariance matrix of ![]() after controlling for
after controlling for ![]() .
.
For an observation with the preceding missing pattern, the conditional distribution of ![]() given
given ![]() is a multivariate normal distribution with the mean vector
is a multivariate normal distribution with the mean vector
|
|
and the conditional covariance matrix
|
|
Bayesian Estimation of the Mean Vector and Covariance Matrix
Suppose that ![]() is an
is an ![]() matrix made up of n
matrix made up of n ![]() independent vectors
independent vectors ![]() , each of which has a multivariate normal distribution with mean zero and covariance matrix
, each of which has a multivariate normal distribution with mean zero and covariance matrix ![]() . Then the SSCP matrix
. Then the SSCP matrix
|
|
has a Wishart distribution ![]() .
.
When each observation ![]() is distributed with a multivariate normal distribution with an unknown mean
is distributed with a multivariate normal distribution with an unknown mean ![]() , then the CSSCP matrix
, then the CSSCP matrix
|
|
has a Wishart distribution ![]() .
.
If ![]() has a Wishart distribution
has a Wishart distribution ![]() , then
, then ![]() has an inverted Wishart distribution
has an inverted Wishart distribution ![]() , where n is the degrees of freedom and
, where n is the degrees of freedom and ![]() is the precision matrix (Anderson, 1984).
is the precision matrix (Anderson, 1984).
Note that, instead of using the parameter ![]() for the inverted Wishart distribution, Schafer (1997) uses the parameter
for the inverted Wishart distribution, Schafer (1997) uses the parameter ![]() .
.
Suppose that each observation in the data matrix ![]() has a multivariate normal distribution with mean
has a multivariate normal distribution with mean ![]() and covariance matrix
and covariance matrix ![]() . Then with a prior inverted Wishart distribution for
. Then with a prior inverted Wishart distribution for ![]() and a prior normal distribution for
and a prior normal distribution for ![]()
|
|
|
|
|
|
|
|
where ![]() is a fixed number.
is a fixed number.
The posterior distribution (Anderson 1984, p. 270; Schafer 1997, p. 152) is
|
|
|
|
|
|
|
|
where ![]() is the CSSCP matrix.
is the CSSCP matrix.
Posterior Step
In each iteration, the posterior step simulates the posterior population mean vector ![]() and covariance matrix
and covariance matrix ![]() from prior information for
from prior information for ![]() and
and ![]() , and the complete sample estimates.
, and the complete sample estimates.
You can specify the prior parameter information by using one of the following methods:
-
PRIOR=JEFFREYS, which uses a noninformative prior
-
PRIOR=INPUT=, which provides a prior information for
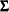 in the data set. Optionally, it also provides a prior information for
in the data set. Optionally, it also provides a prior information for 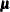 in the data set.
in the data set.
-
PRIOR=RIDGE=, which uses a ridge prior
The next four subsections provide details of the posterior step for different prior distributions.
1. A Noninformative Prior
Without prior information about the mean and covariance estimates, you can use a noninformative prior by specifying the PRIOR=JEFFREYS option. The posterior distributions (Schafer, 1997, p. 154) are
|
|
|
|
|
|
|
|
2. An Informative Prior for 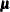 and
and 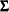
When prior information is available for the parameters ![]() and
and ![]() , you can provide it with a SAS data set that you specify with the PRIOR=INPUT= option:
, you can provide it with a SAS data set that you specify with the PRIOR=INPUT= option:
|
|
|
|
|
|
|
|
To obtain the prior distribution for ![]() , PROC MI reads the matrix
, PROC MI reads the matrix ![]() from observations in the data set with
from observations in the data set with _TYPE_=‘COV’, and it reads ![]() from observations with
from observations with _TYPE_=‘N’.
To obtain the prior distribution for ![]() , PROC MI reads the mean vector
, PROC MI reads the mean vector ![]() from observations with
from observations with _TYPE_=‘MEAN’, and it reads ![]() from observations with
from observations with _TYPE_=‘N_MEAN’. When there are no observations with _TYPE_=‘N_MEAN’, PROC MI reads ![]() from observations with
from observations with _TYPE_=‘N’.
The resulting posterior distribution, as described in the section Bayesian Estimation of the Mean Vector and Covariance Matrix, is given by
|
|
|
|
|
|
|
|
where
|
|
3. An Informative Prior for 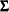
When the sample covariance matrix ![]() is singular or near singular, prior information about
is singular or near singular, prior information about ![]() can also be used without prior information about
can also be used without prior information about ![]() to stabilize the inference about
to stabilize the inference about ![]() . You can provide it with a SAS data set that you specify with the PRIOR=INPUT= option.
. You can provide it with a SAS data set that you specify with the PRIOR=INPUT= option.
To obtain the prior distribution for ![]() , PROC MI reads the matrix
, PROC MI reads the matrix ![]() from observations in the data set with
from observations in the data set with _TYPE_=‘COV’, and it reads ![]() from observations with
from observations with _TYPE_=‘N’.
The resulting posterior distribution for ![]() (Schafer, 1997, p. 156) is
(Schafer, 1997, p. 156) is
|
|
|
|
|
|
|
|
Note that if the PRIOR=INPUT= data set also contains observations with _TYPE_=‘MEAN’, then a complete informative prior for both ![]() and
and ![]() will be used.
will be used.
4. A Ridge Prior
A special case of the preceding adjustment is a ridge prior with ![]() = Diag
= Diag ![]() (Schafer, 1997, p. 156). That is,
(Schafer, 1997, p. 156). That is, ![]() is a diagonal matrix with diagonal elements equal to the corresponding elements in
is a diagonal matrix with diagonal elements equal to the corresponding elements in ![]() .
.
You can request a ridge prior by using the PRIOR=RIDGE= option. You can explicitly specify the number ![]() in the PRIOR=RIDGE=
in the PRIOR=RIDGE=![]() option. Or you can implicitly specify the number by specifying the proportion p in the PRIOR=RIDGE=p option to request
option. Or you can implicitly specify the number by specifying the proportion p in the PRIOR=RIDGE=p option to request ![]() .
.
The posterior is then given by
|
|
|
|
|
|
|
|